WGCNA scale-free测试
只有三个样本,其实根本不适合做WGCNA。即便按照流程分析完,得到的数据结果也不理想,network根本不可能scale free。
利用WGCNA自带的femaleliver的数据,随机抽取其中三个样本进行softpower的计算,重复5次。
# Load the WGCNA package
library(WGCNA);
# The following setting is important, do not omit.
options(stringsAsFactors = FALSE);
#Read in the female liver data set
femData = read.csv("LiverFemale3600.csv");
datExpr0 = as.data.frame(t(femData[, -c(1:8)]));
names(datExpr0) = femData$substanceBXH;
rownames(datExpr0) = names(femData)[-c(1:8)];
gsg = goodSamplesGenes(datExpr0, verbose = 3);
gsg$allOK
if (!gsg$allOK)
{
# Optionally, print the gene and sample names that were removed:
if (sum(!gsg$goodGenes)>0)
printFlush(paste("Removing genes:", paste(names(datExpr0)[!gsg$goodGenes], collapse = ", ")));
if (sum(!gsg$goodSamples)>0)
printFlush(paste("Removing samples:", paste(rownames(datExpr0)[!gsg$goodSamples], collapse = ", ")));
# Remove the offending genes and samples from the data:
datExpr0 = datExpr0[gsg$goodSamples, gsg$goodGenes]
}
dim(datExpr0)
nGenes = ncol(datExpr0)
nSamples = nrow(datExpr0)
set.seed(44)
for (i in 1:5){
# randomly pick up 3 samples
random_n=sample(length(nSamples),3)
datExpr1=datExpr0[random_n,]
# Choose a set of soft-thresholding powers
powers = c(c(1:10), seq(from = 12, to=20, by=2))
# Call the network topology analysis function
sft = pickSoftThreshold(datExpr1, powerVector = powers, verbose = 5)
# Plot the results:
pdf(paste0("try",i,"_sft.pdf"))
par(mfrow = c(1,2));
cex1 = 0.9;
# Scale-free topology fit index as a function of the soft-thresholding power
plot(sft$fitIndices[,1], -sign(sft$fitIndices[,3])*sft$fitIndices[,2],
xlab="Soft Threshold (power)",ylab="Scale Free Topology Model Fit,signed R^2",type="n",
main = paste("Scale independence"));
text(sft$fitIndices[,1], -sign(sft$fitIndices[,3])*sft$fitIndices[,2],
labels=powers,cex=cex1,col="red");
# this line corresponds to using an R^2 cut-off of h
abline(h=0.90,col="red")
# Mean connectivity as a function of the soft-thresholding power
plot(sft$fitIndices[,1], sft$fitIndices[,5],
xlab="Soft Threshold (power)",ylab="Mean Connectivity", type="n",
main = paste("Mean connectivity"))
text(sft$fitIndices[,1], sft$fitIndices[,5], labels=powers, cex=cex1,col="red")
dev.off()
}
使用全部样本(135个),得到的softpower结果如下,
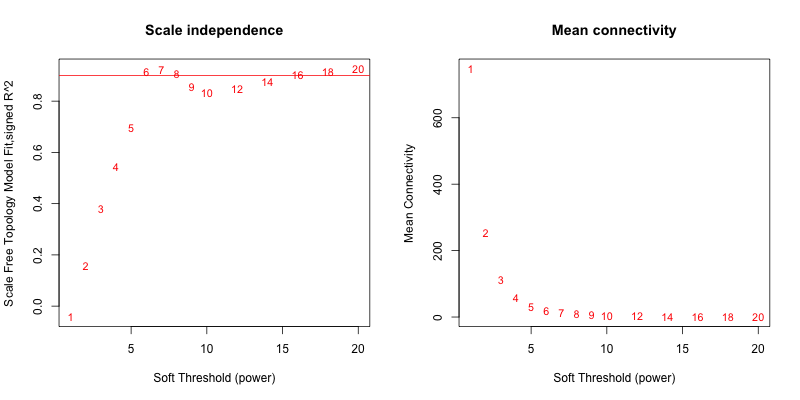
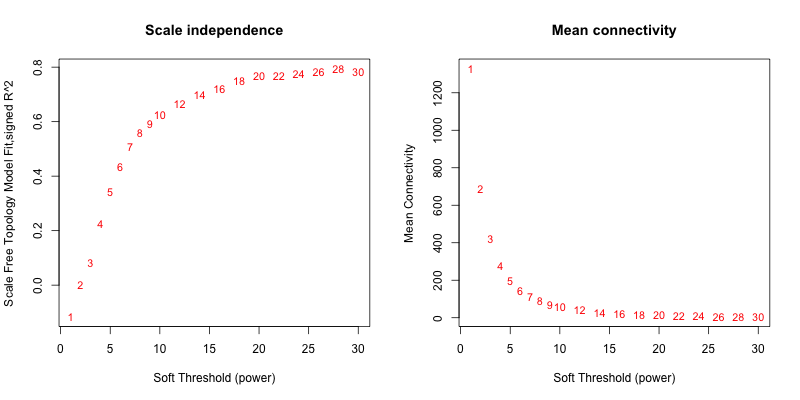
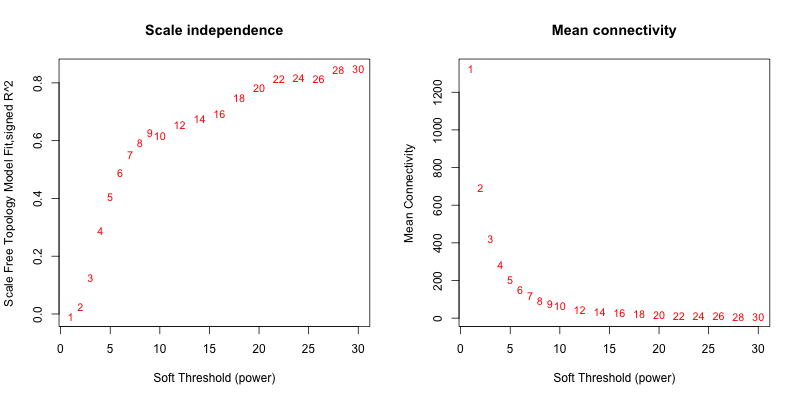
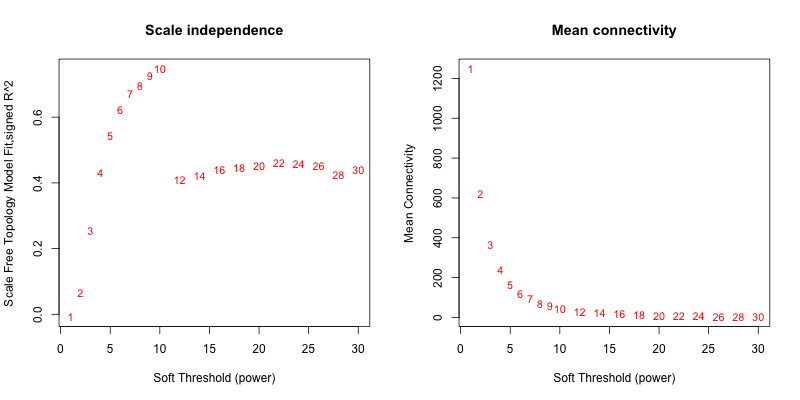
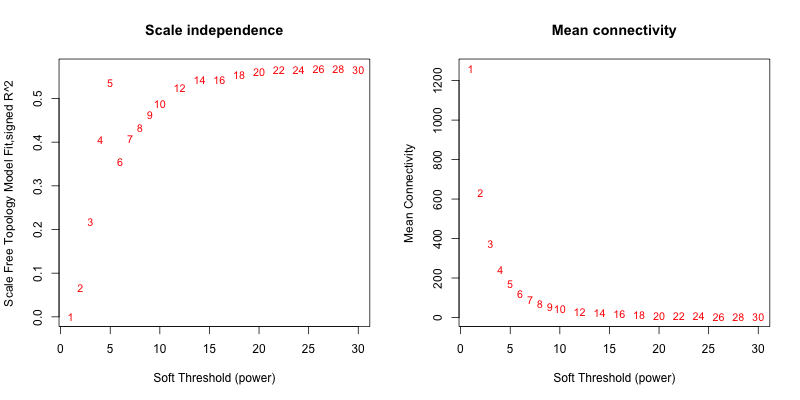
5次随机抽取3个样本,得到的softpower结果如下
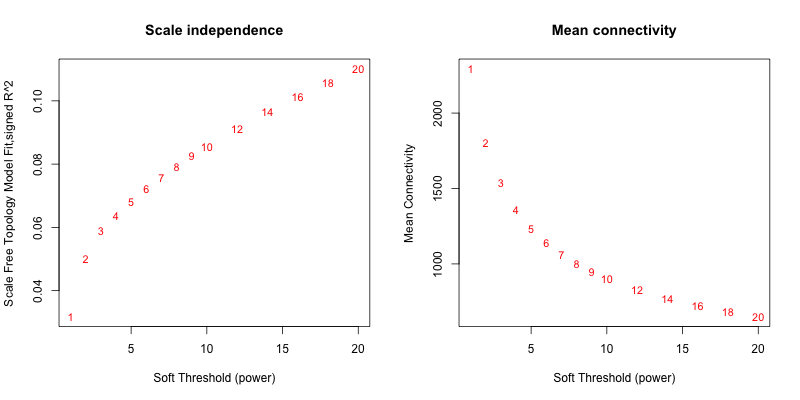
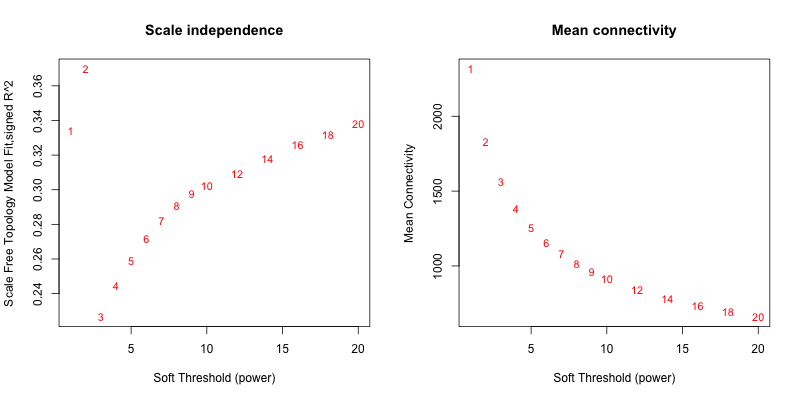
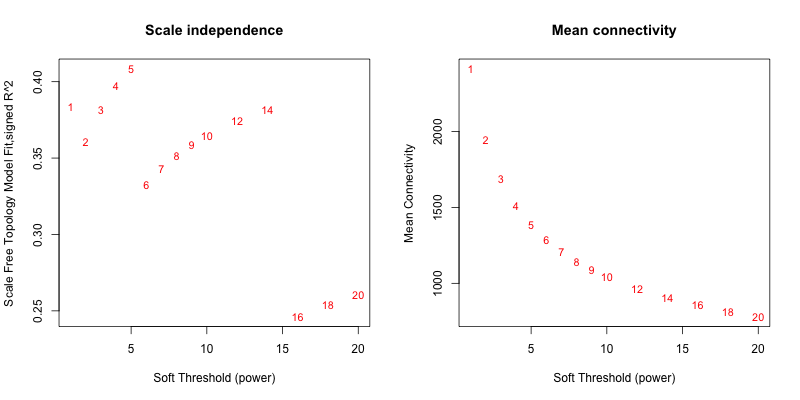
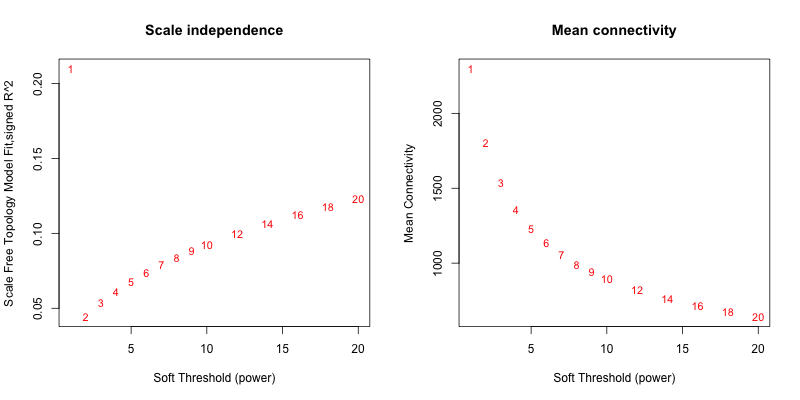
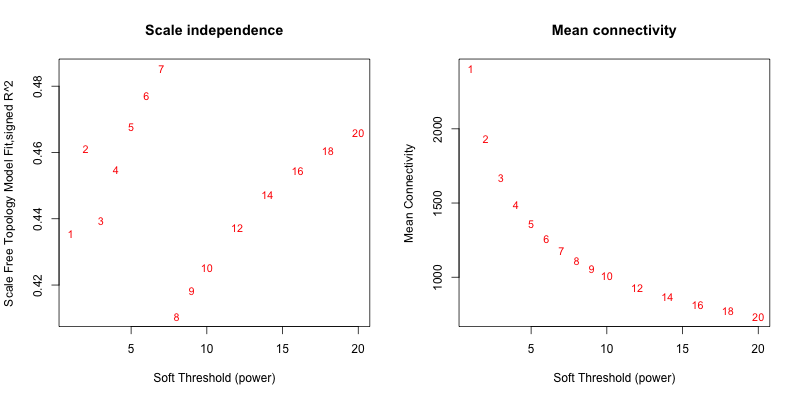
5次随机抽取6个样本,得到的softpower结果如下
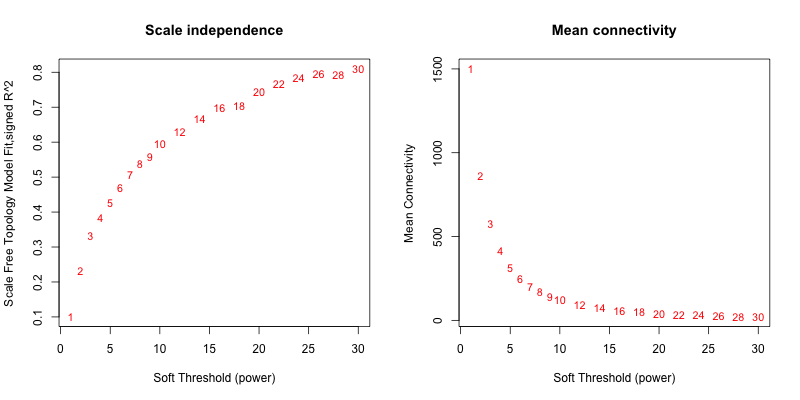
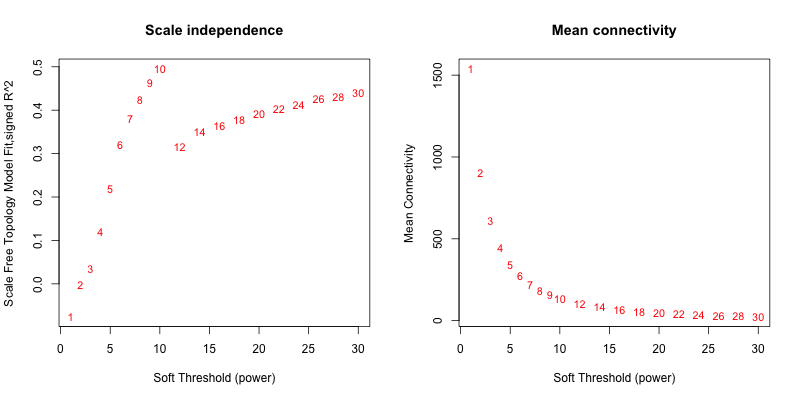
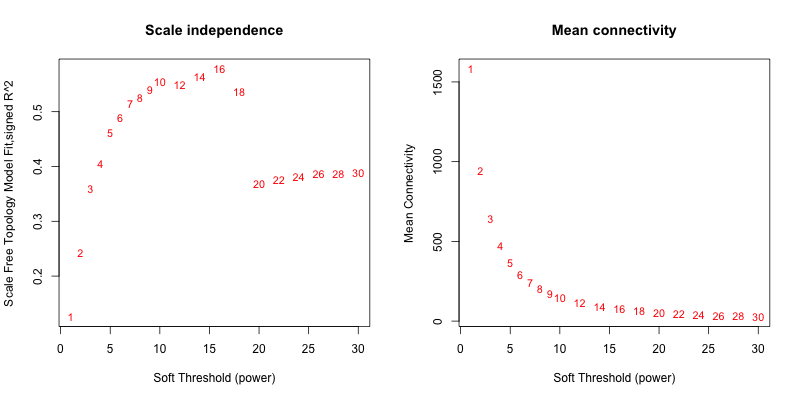
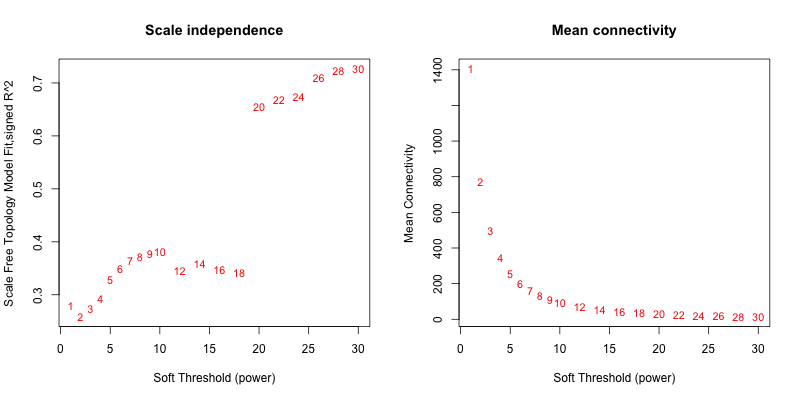
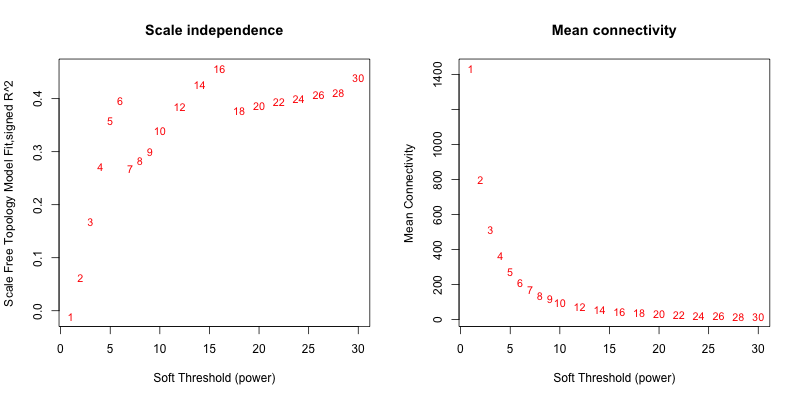
5次随机抽取9个样本,得到的softpower结果如下
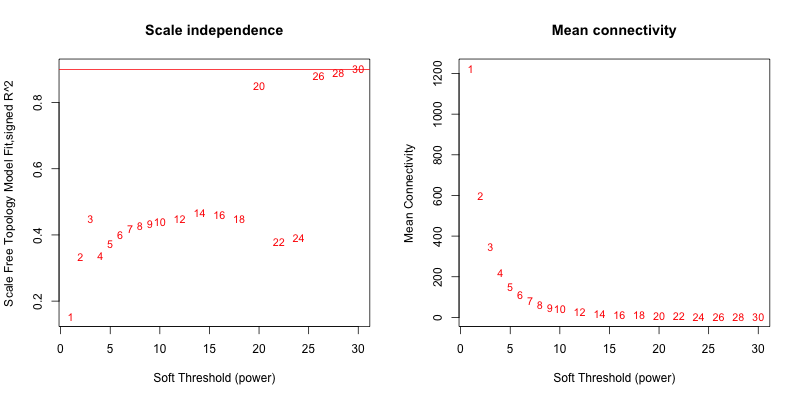
5次随机抽取12个样本,得到的softpower结果如下
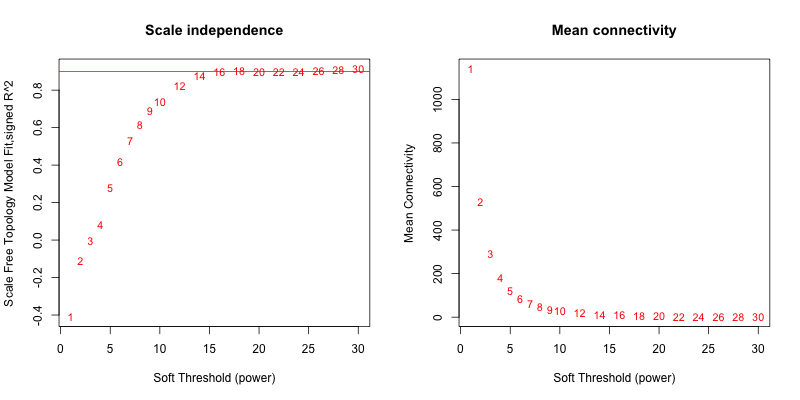
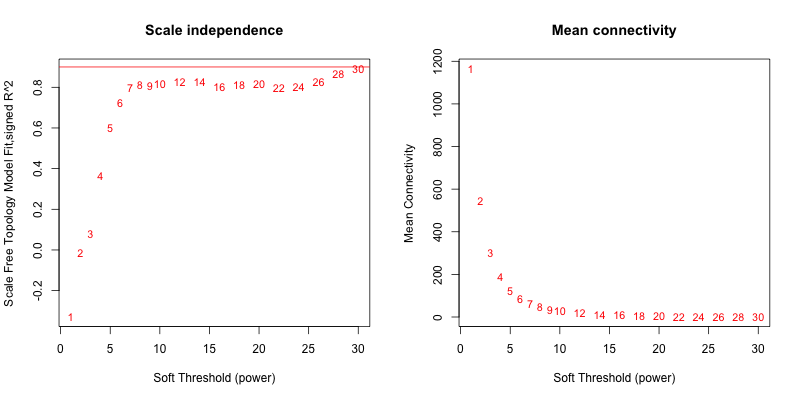
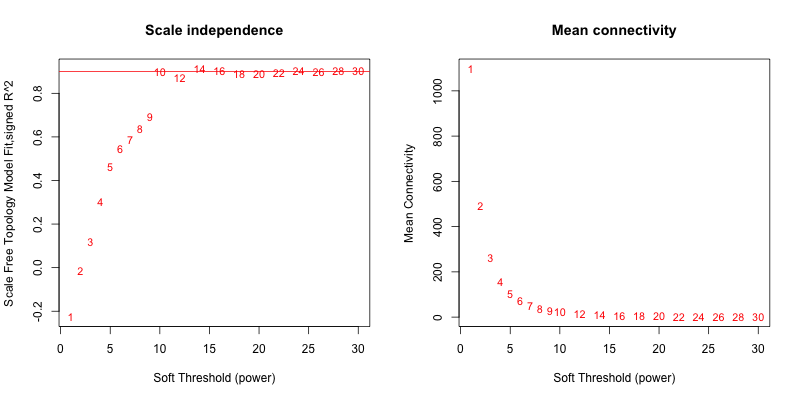
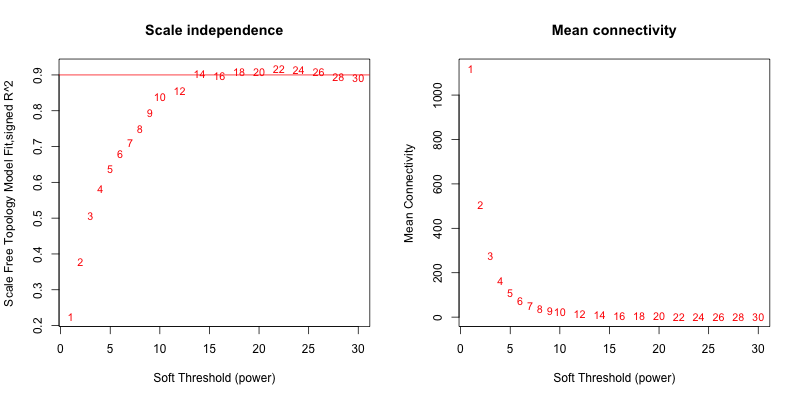
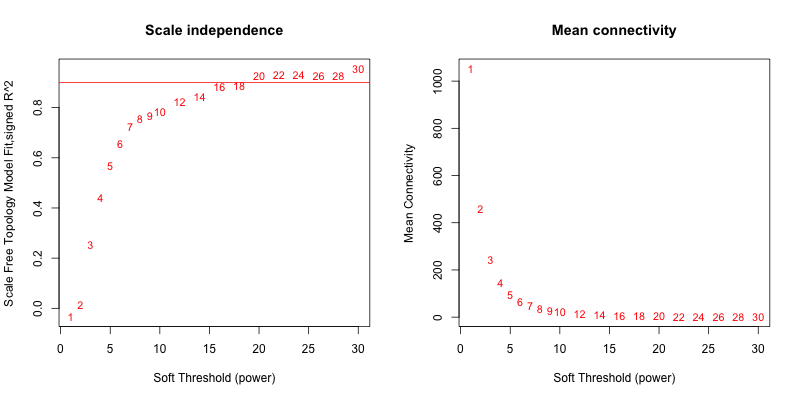
可以看到,参与计算的样本数越多,得到的softpower结果越容易达到scale-free。